Bioinformatics: an overview of WSLH activities
Kelsey Florek, PhD, MPH Senior Genomics and Data Scientist Wisconsin State Laboratory of Hygiene May 4, 2022

What is bioinformatics?
"The branch of science concerned with information and information flow in biological systems, especially the use of computational methods in genetics and genomics.” - Oxford English Dictionary
What is bioinformatics?
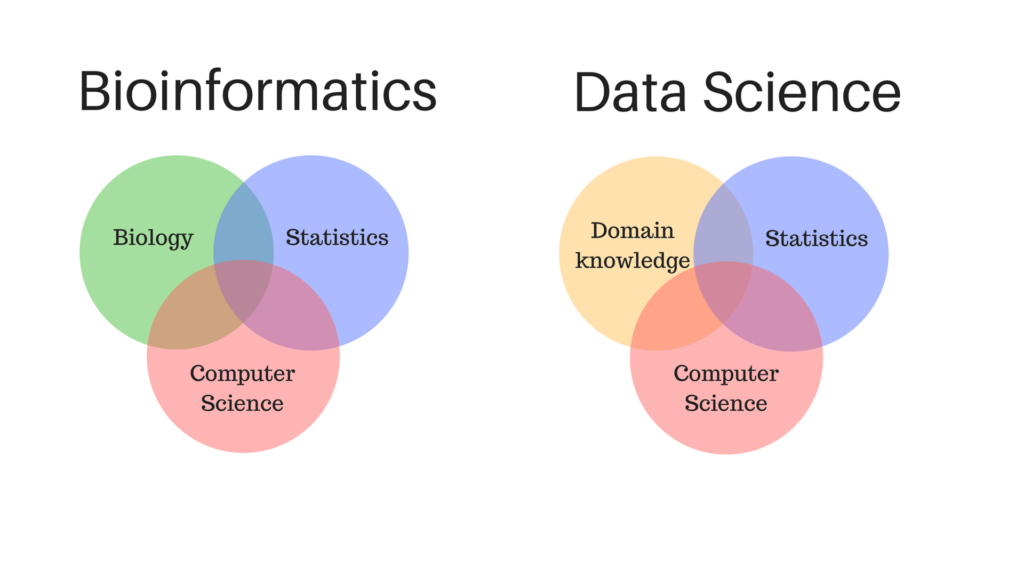
Applying Bioinformatics to Public Health (Genomic Epidemiology)
Turning a Sample into Genomic Data

Increases in data requires advanced analyses (MiSeq)
- 15,000,000,000 ATGC's generated per sequencing run
- 40,000 - 150,000 words in a novel
- average word length in English is 4.79
- one sequencing run would generate 32,963 novels with 95,000 words each
Increases in data requires advanced analyses (Nextseq 2000)
- 360,000,000,000 ATGC's generated per sequencing run
- 40,000 - 150,000 words in a novel
- average word length in English is 4.79
- one sequencing run would generate 791,121 novels with 95,000 words each
Infectious Disease Genomics at WSLH
Pathogens we are currently sequencing
- Influenza
- SARS-CoV-2
- Salmonella
- E. coli
- Shigella
- Cyclospora cayetanensis
- Campylobacter
- Listeria monocytogenes
- Vibrio cholerae
- Vibrio parahaemolyticus
- Cronobacter
- Enterobacteriaceae
- Acinetobacter baumannii
Pathogens we are planning to sequence
- Cryptosporidium
- Mycobacterium tuberculosis
- Mycobacteria
- Candida auris
- Hepatitus C Virus
- Metagenomics
Application of Bioinformatics for SARS-CoV-2
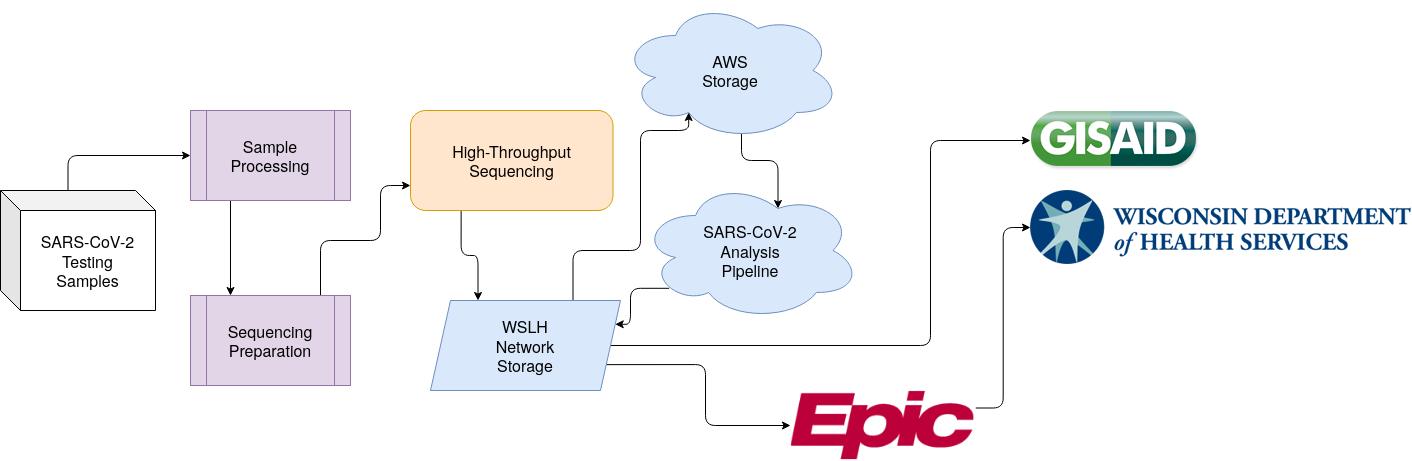
Application of Bioinformatics for SARS-CoV-2
Cloud Computing
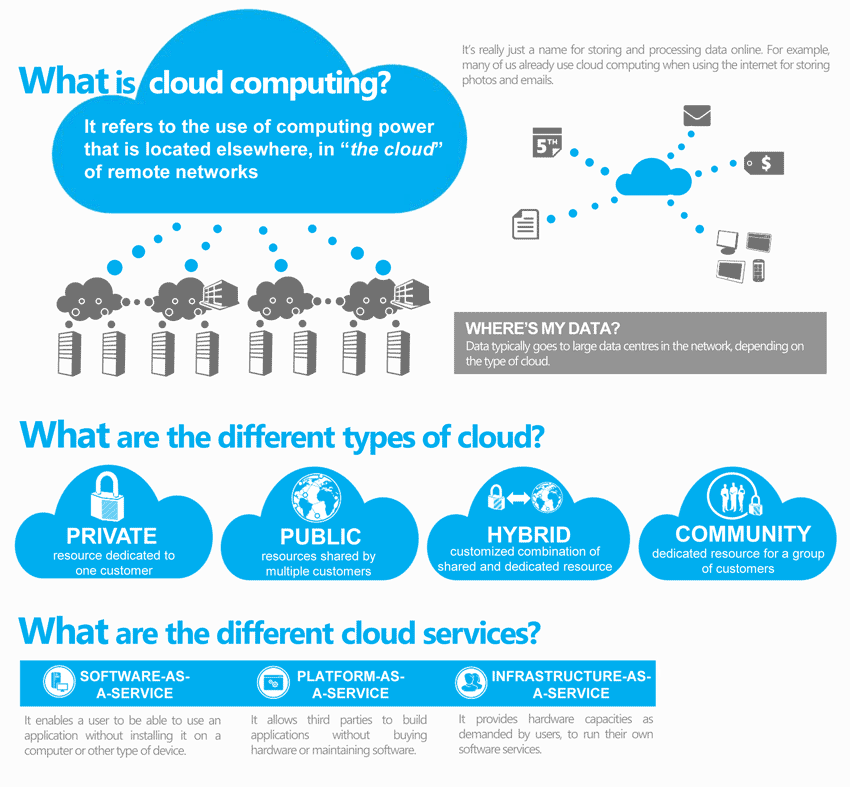
Why cloud computing is valuable for Bioinformatics?
- Cost Efficient Data Storage: ~$20/Month for 1,000 GB
- Cost Efficient Compute: Running highly complex analytical workflows cost on average $1-2 an hour
- Extreme scalability: With no intervention we can shift from 20 samples a week to over 10,000s
- Pay for what we use: Cost is only incurred when resources are used, this allows us to keep our workflows cheap and efficient
Why cloud computing is valuable for Bioinformatics?
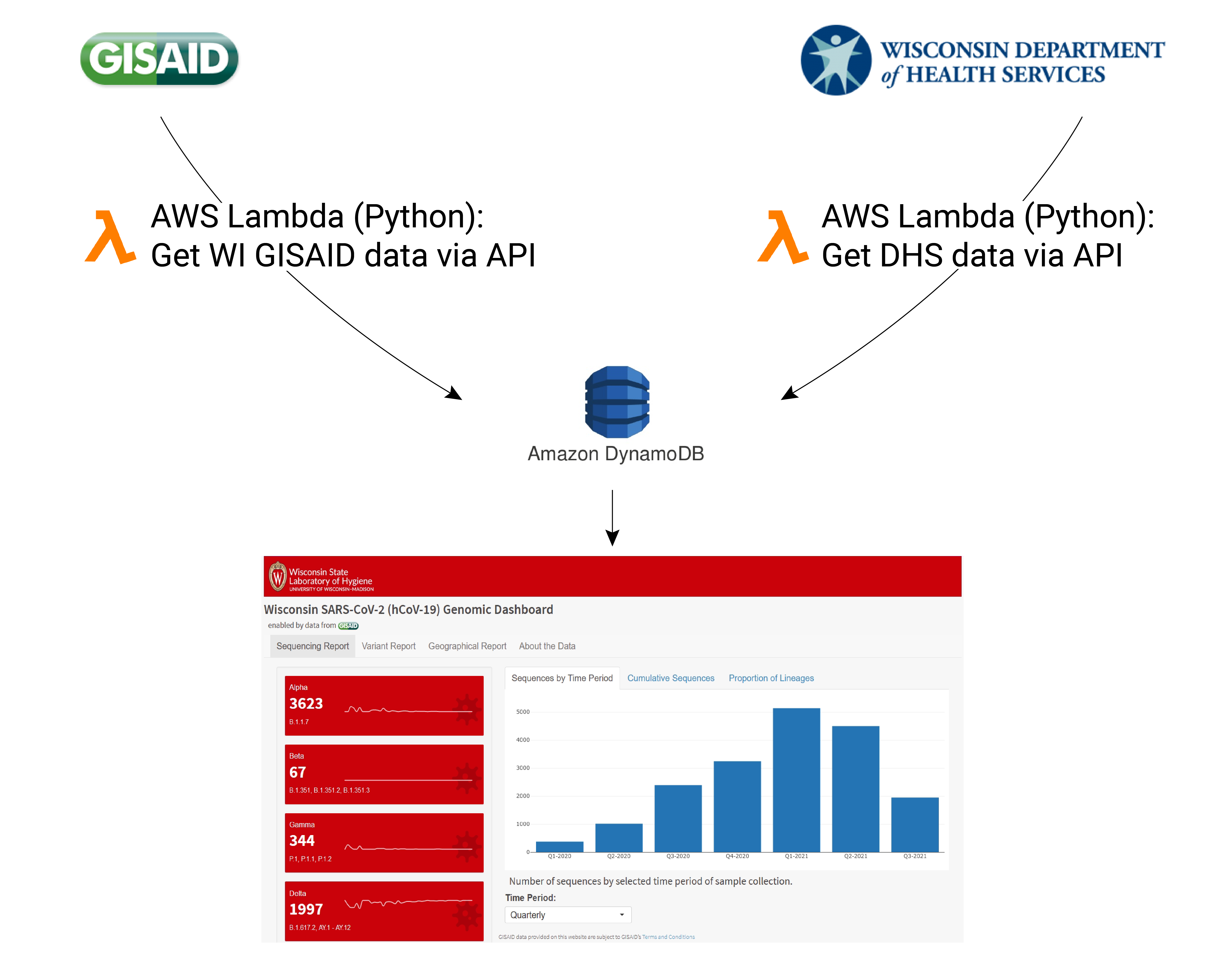
Connecting data within WSLH
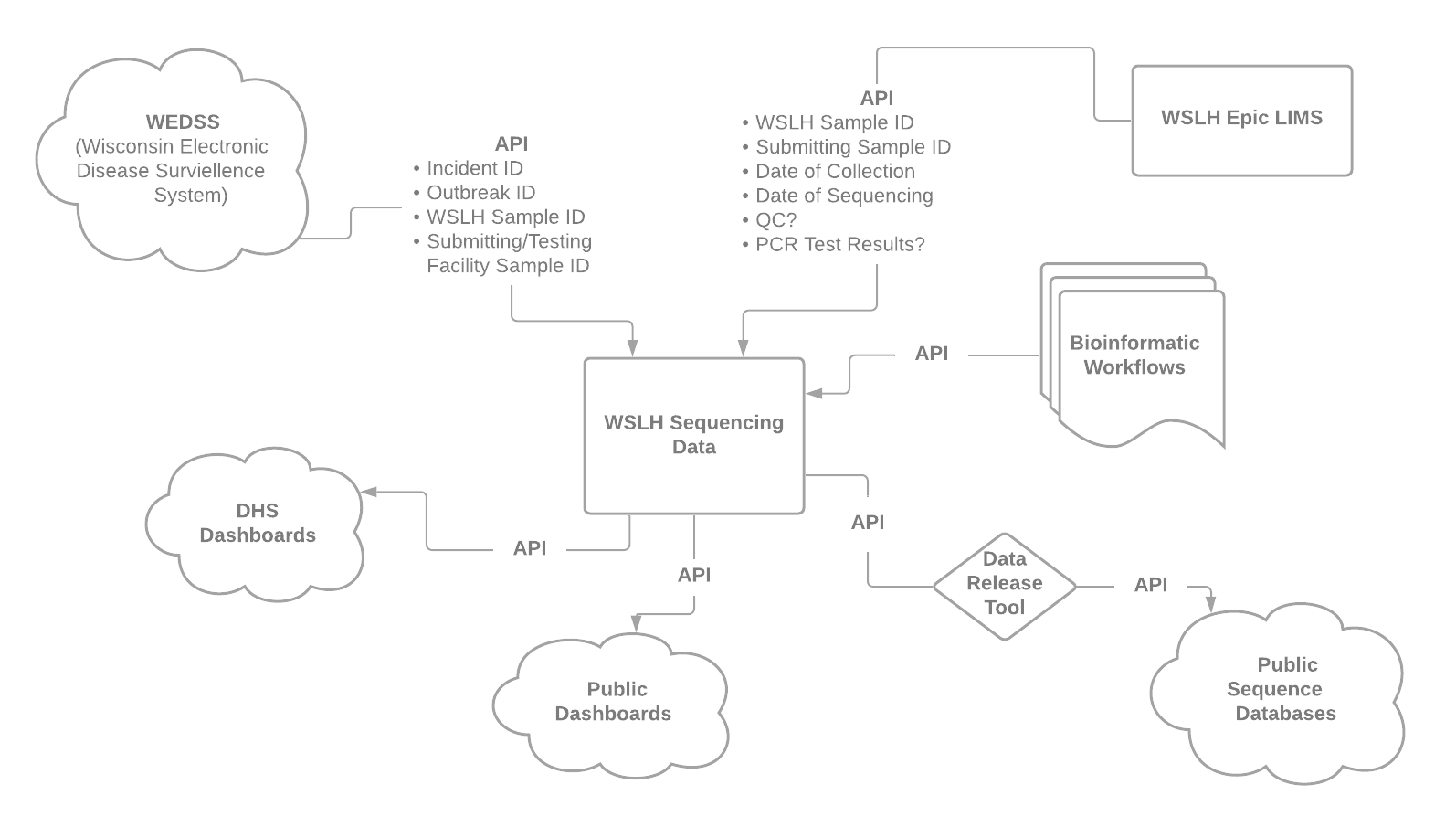
Connecting data within WSLH
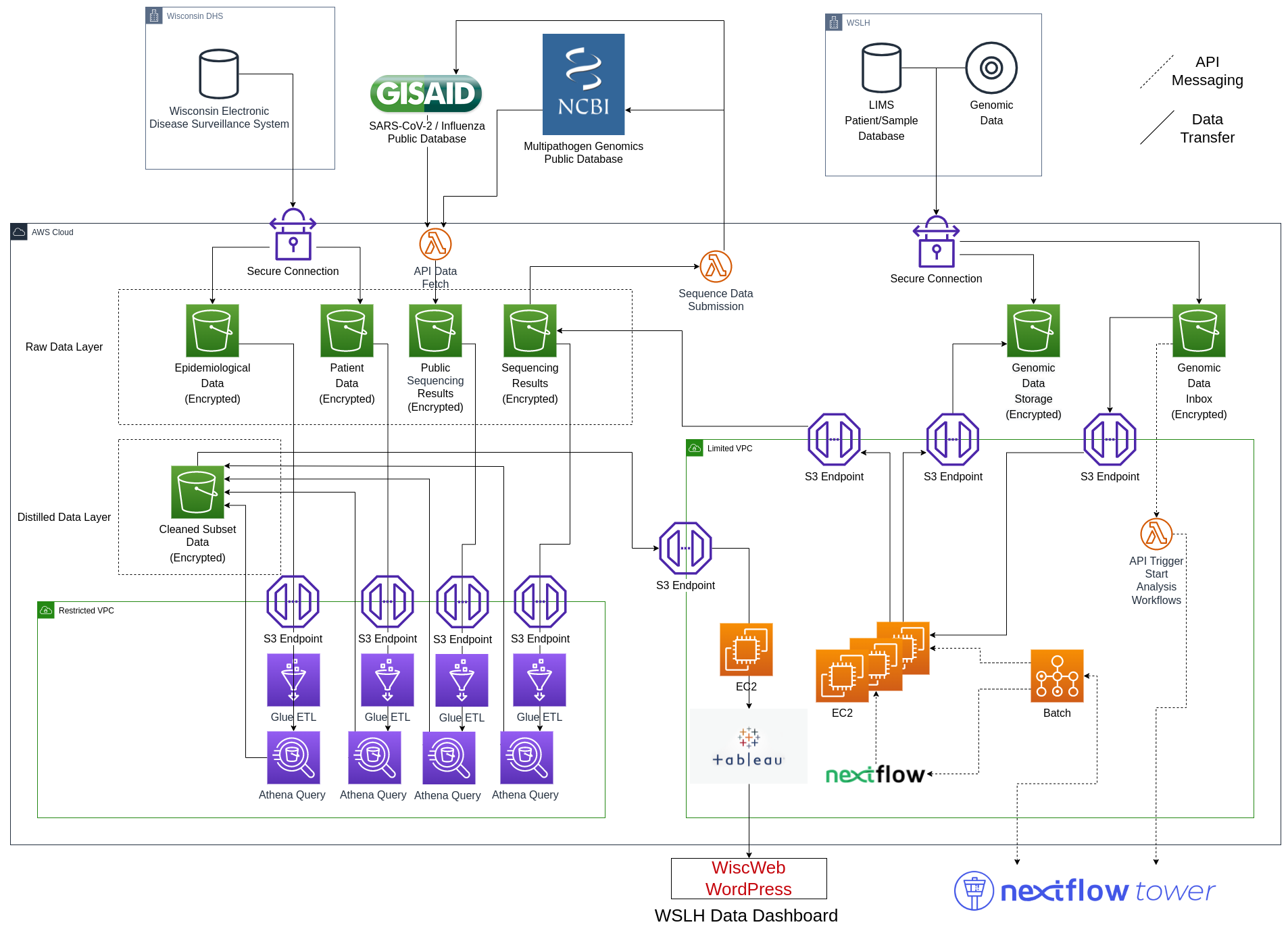
Where we are heading
WSLH is a leading Public Health Lab in Infectious Disease Genomic Epidemiology
- CDC Bioinformatics Regional Resource for the Midwest
- Member of PHA4GE: Public Health Alliance for Genomic Epidemiology
- Member of Spheres: SARS-CoV-2 Sequencing for Public Health Emergency Response, Epidemiology and Surveillance
- Member of StaPH-B: State Public Health Bioinformatics Workgroup
- Currently Applying to become CDC Genomic Center of Excellence
WSLH is innovating a new application of genomic epidemiology
- WSLH is the only laboratory that augments genomic analytics with Cloud technology
- University Support
- Expertise
- Funding
- The future of Public Health is in data
- Precision Public Health: the right intervention to the right population at the right time
- genomics, spacial data, epidemiology, data linkage, and predictive analytics
What we need to move forward successfully in the field of Public Health Bioinformatics
- Understanding - Our lab space is our computers, as one might need equipment in a lab we need software and tools
- Flexibility - Our approach is unlike anything that has existed in the past, our approach, tooling, and methods change very frequently
- Support - We are innovating and navigating in a space that is new to everyone, we need support to make our path less treacherous