Advancing Data Strategies for Tracking Infectious Diseases
Kelsey Florek, PhD, MPH Senior Genomics and Data Scientist Wisconsin State Laboratory of Hygiene June 6, 2024
Slides live at:
www.k-florek.net/talks
www.k-florek.net/talks

Next Generation Sequencing for Infectious Disease Surveillance
What is Next Generation Sequencing?
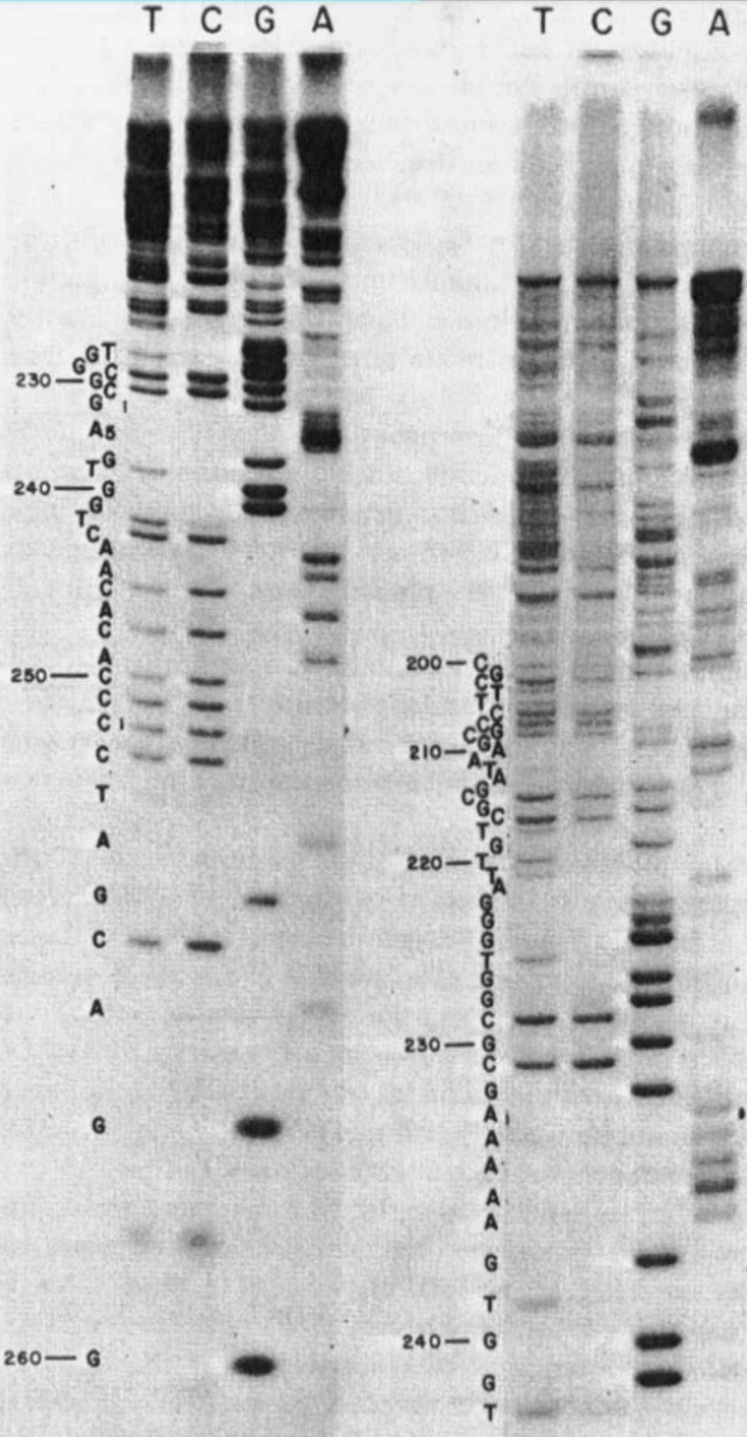
Sanger Sequencing
Next Generation Sequencing (Illumina)

High-throughput requires advanced analytics (Nextseq 2000)
- 30,000,000,000 ATGC's generated per sequencing run
- 40,000 - 150,000 words in a novel
- average word length in English is 4.79
- one sequencing run would generate 62,000 novels with 100,000 words each
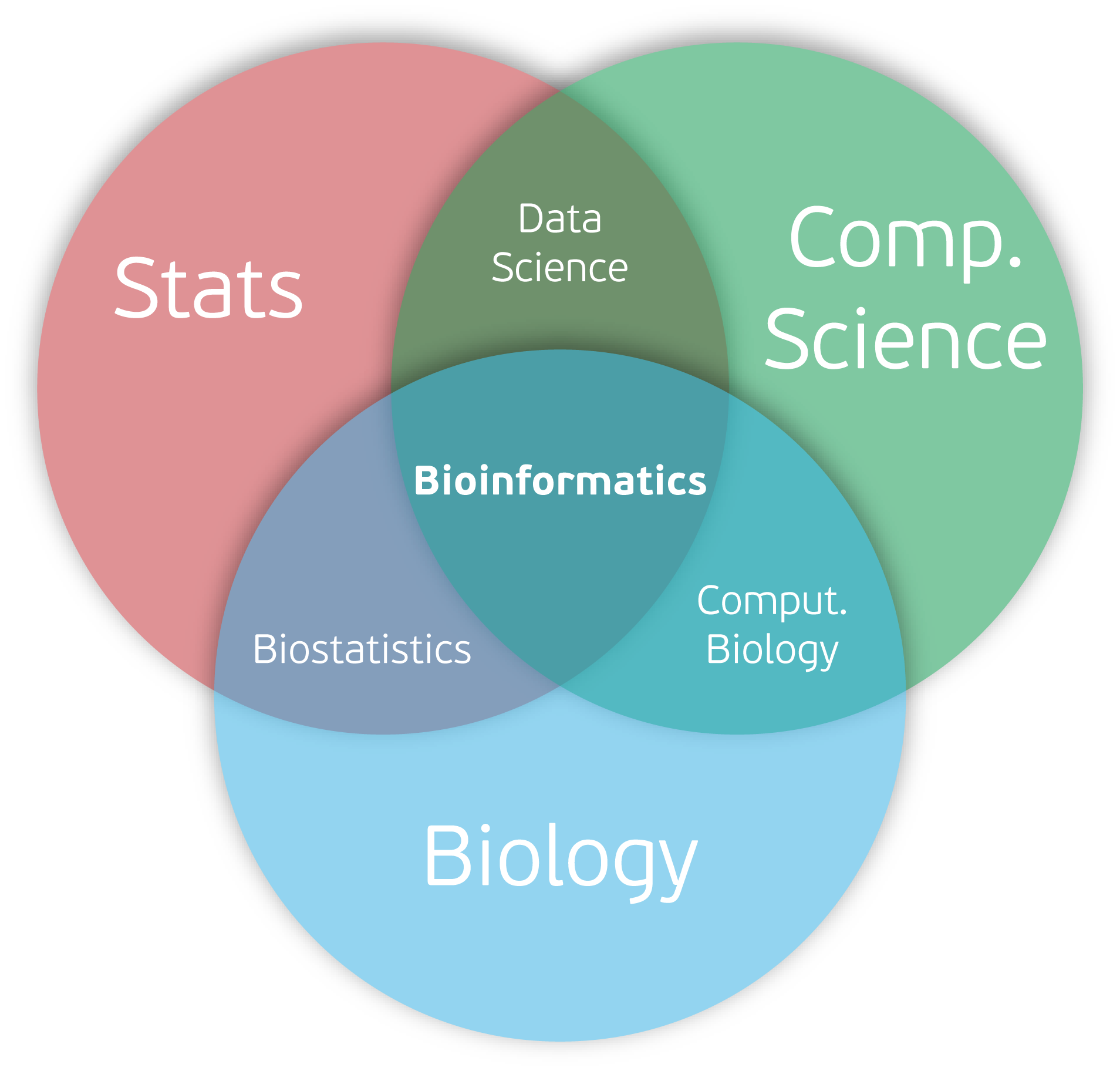
Anatomy of a Bioinformatics Workflow

include { SAMPLESHEET_CHECK } from '../../modules/local/samplesheet_check'
workflow INPUT_CHECK {
take:
samplesheet // file: /path/to/samplesheet.csv
main:
SAMPLESHEET_CHECK ( samplesheet )
.csv
.splitCsv ( header:true, sep:',' )
.map { create_fasta_channel(it) }
.set { reads }
emit:
// channel: [ val(meta), [ path_to_reads ] ]
reads
// channel: [ samplesheet.valid.csv ]
csv = SAMPLESHEET_CHECK.out.csv
// channel: [ versions.yml ]
versions = SAMPLESHEET_CHECK.out.versions
}
// Function to get list of [ meta, [ fasta ] ]
def create_fasta_channel(LinkedHashMap row) {
// create meta map
def meta = [:]
meta.id = row.sample
def fasta_meta = []
if (!file(row.fasta).exists()) {
exit 1
} else {
fasta_meta = [ meta, [ file(row.fasta) ] ]
}
return fasta_meta
}
Sequencing Reads
@M05192:295:000000000-K6N36:1:1101:9502:1212 1:N:0:NAGCGCTC+NCGTAAGA
GCGTTACATGAGGCTTATACTGAAACATTGCCTAATCCCGCCCGGTGGAAAGCTAAAAANTCCTNTGAACTGCNGGGCTATTCAGAAGNNNN
+
CCCCCGGGGGGGGGGGGGGGGGGGGGGGGGGGGFF@@FGGGGGGGGGGGGGGDGGGGFG#:C@F#:@FGGGGG#:C@FFFGGGAFGF?####
@M05192:295:000000000-K6N36:1:1101:13064:1213 1:N:0:NAGCGCTC+NCGTAAGA
TAGTGGCACTGTTTGACCATCAGCAACGCATTGGTGAACTGATGCCGGAGCGGCGTTTTNACNANGCACGTCGNCAGCAAATGATGGANNNNNNNNNNNN
+
CCCCCGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG#:D#:#6CFGGGGG#:DFGGGFGG??FGG############
@M05192:295:000000000-K6N36:1:1101:18301:1216 1:N:0:NAGCGCTC+NCGTAAGA
GCCCGGTGGTGTAATTTGCGCCCTCCGAACAAAGCCACGCCACCAGGCTGGCAATCTCANACNTNGCGCCAAANCGCCGCAGAGGAATNNNNNNNN
+
CCCCCGGGGGFGGGGGGGGGGCFCGGGGGGGGGFGGGGGDGGEGGFGGGGGGGGGGGGF#:C#:#::@D@7FE#6CC#FGGGGCFGFE########
@M05192:295:000000000-K6N36:1:1102:8809:23354 1:N:0:TAGCGCTC+GCGTAAGA
GTGTTATTTGCGTGTCGCGGGCATTATGGCGGAGTACTCTCAGCCTGACGATATGATGGTGGTTTCCGCCGCCGGTAGCACCACTAACCAGTTGAT
+
CCC#CFGGFGGDECG9@FFGGGGFGG#,,C7::@F@FFGFFGGGGGGFGEEF@C,9EA9C@BBFGEF>FGECFGGF+F#C##BDFGFFFFE,??9,
@M05192:295:000000000-K6N36:1:1102:12222:23342 1:N:0:TAGCGCTC+GCGTAAGA
AAGCTAACCGATGCGGATAATGCCGCCGATGGCATTTTTTTCCCCGCCCTTGAGCAAAATATGATGGGTGCGGTGTTAATTAACGAAAATGATGAAGT
+
CCCCCGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGCFGGGGGGGGGGGGFGCEFGGGGGGGGGGGGGGEF
@M05192:295:000000000-K6N36:1:1102:17996:23344 1:N:0:TAGCGCTC+GCGTAAGA
GAGCAGGATAAAACCTACAAAATTACAGTTCTGCATACCAATGATCATCATGGGCATTTTTGGCGCAATGAATATGGCGAATATGGTCTGGCG
+
CCCCCGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGDGGGGGGGGGGGGGGGGGGFGGGGGG
Phylogenetic Tree
WSLH CDD Bioinformatic Workflows

- Spriggan
- AR Report Generator
- SPNTypeID

Public Health Application of
Infectious Disease Genomics
- Pathogen Surveillance
- trends
- prioritization and risk assessment
- early warning system
- Outbreak Response
- source tracking
- containment
- risk assessment
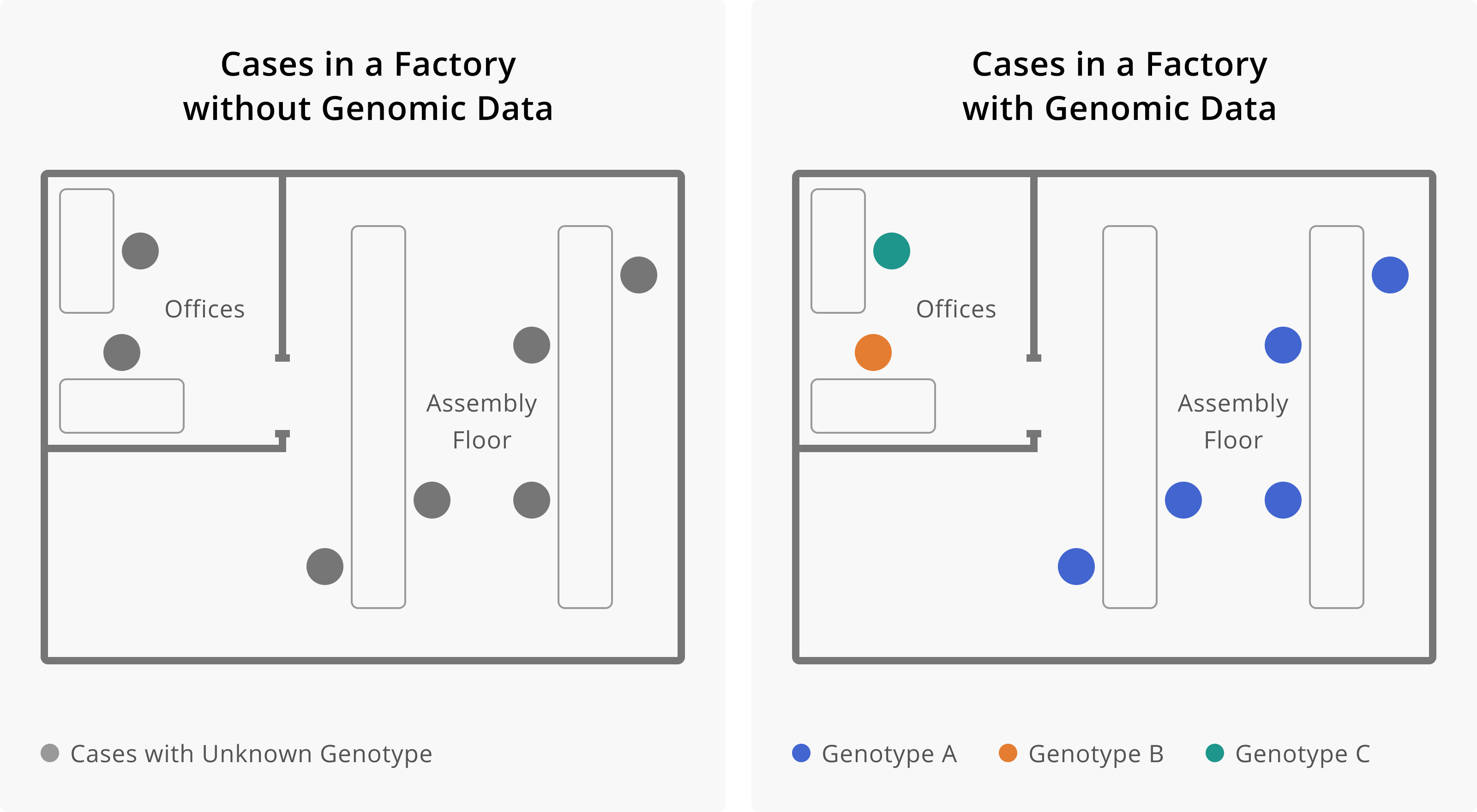
Application of Genomic Data to Outbreaks
Application of Genomic Data to Surveillance
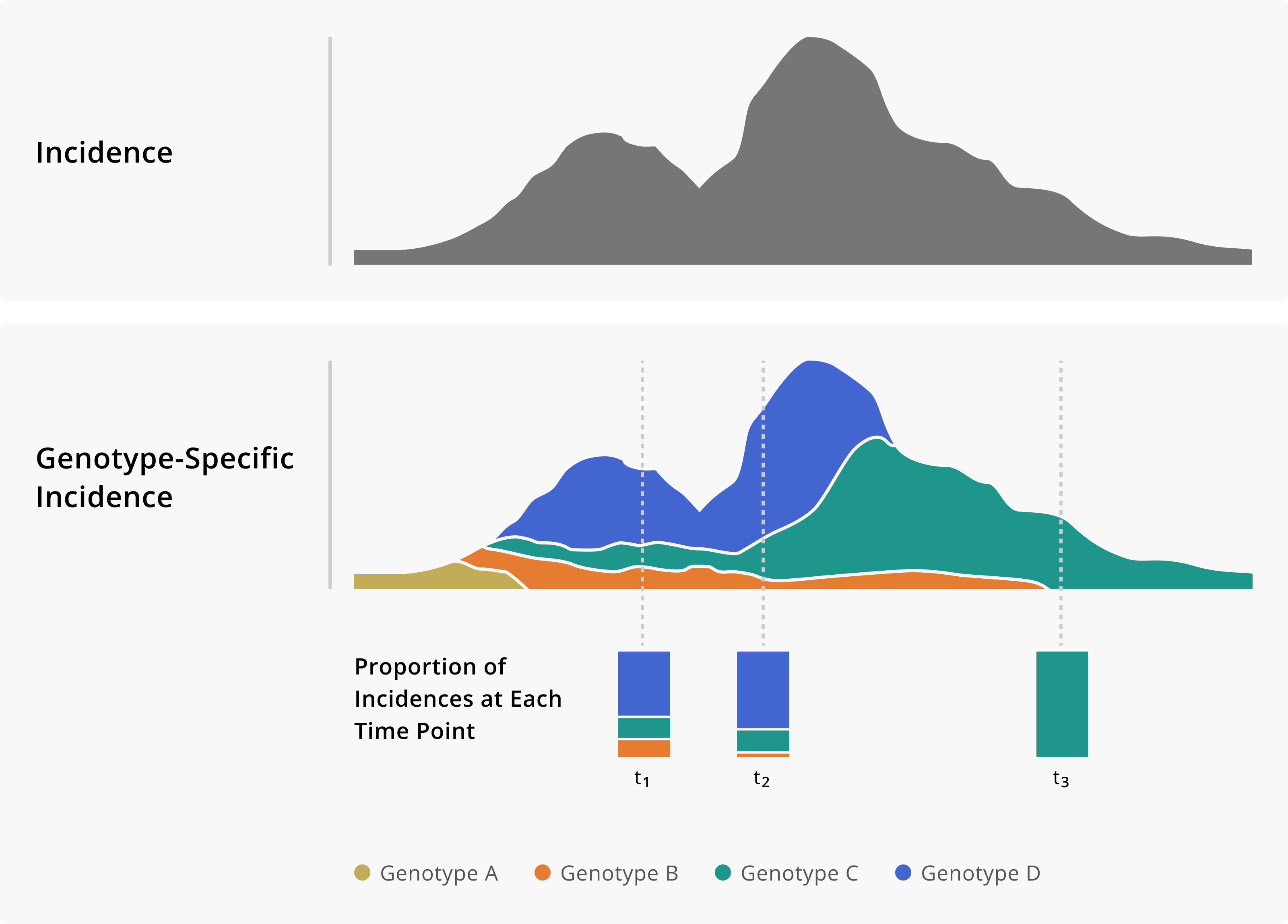
SARS-CoV-2 Variant Waves in WI
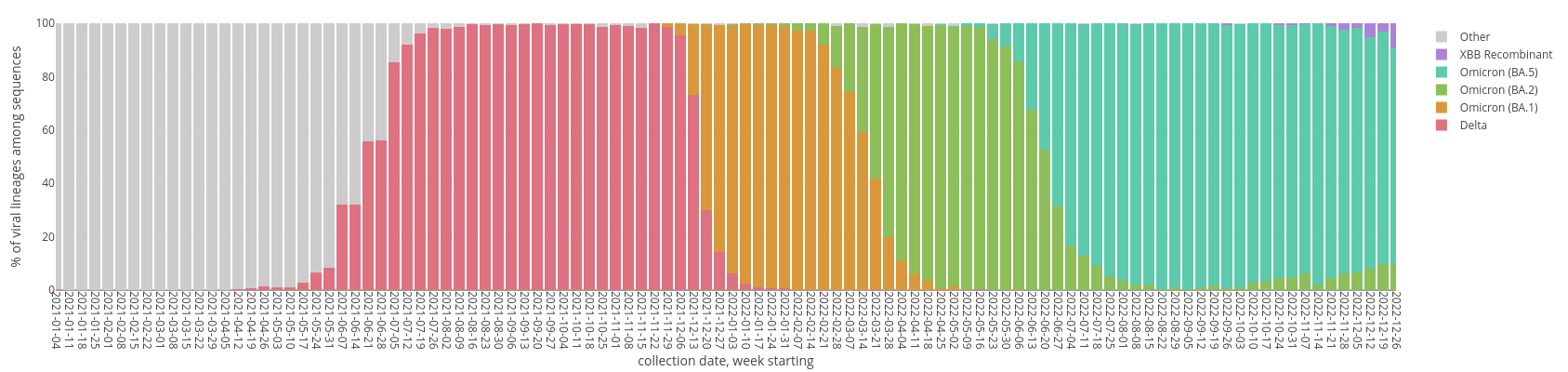
SARS-CoV-2 Variant Waves in WI
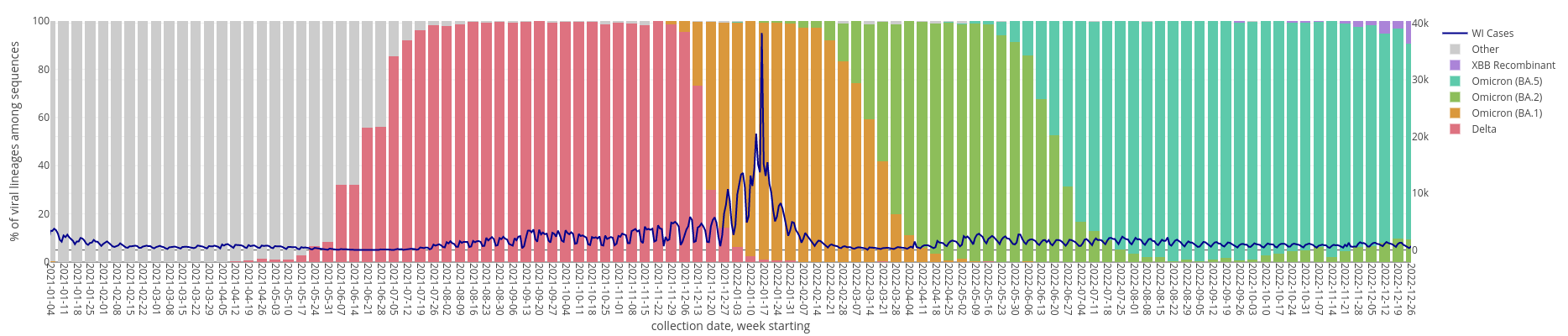
Pathogen Surveillance: SARS-CoV-2
SARS-CoV-2 Wastewater
SARS-CoV-2 Clinical
Advancing Genomics in the Cloud
What is the cloud?
Cloud Data Strategies
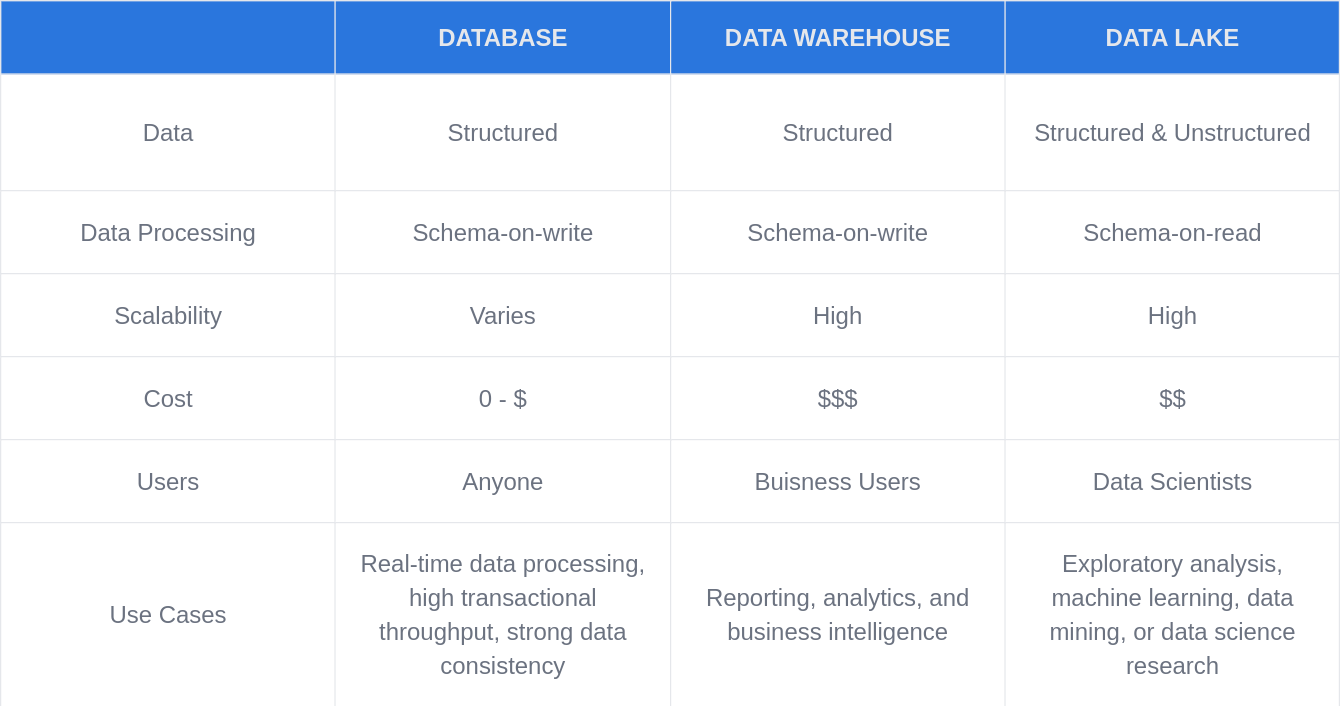
Genomic Data Lake

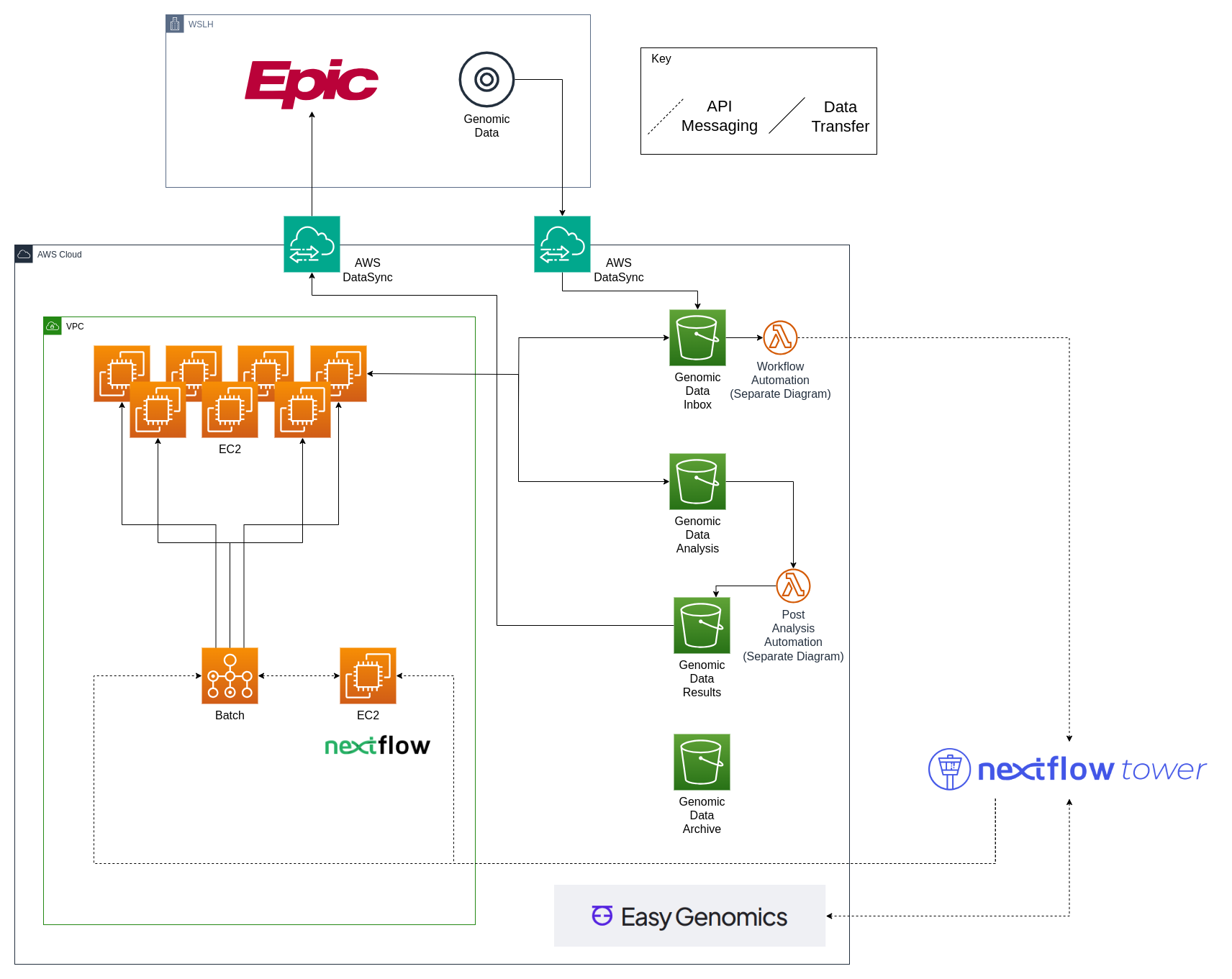
Genomic Data Cloud Infrastructure
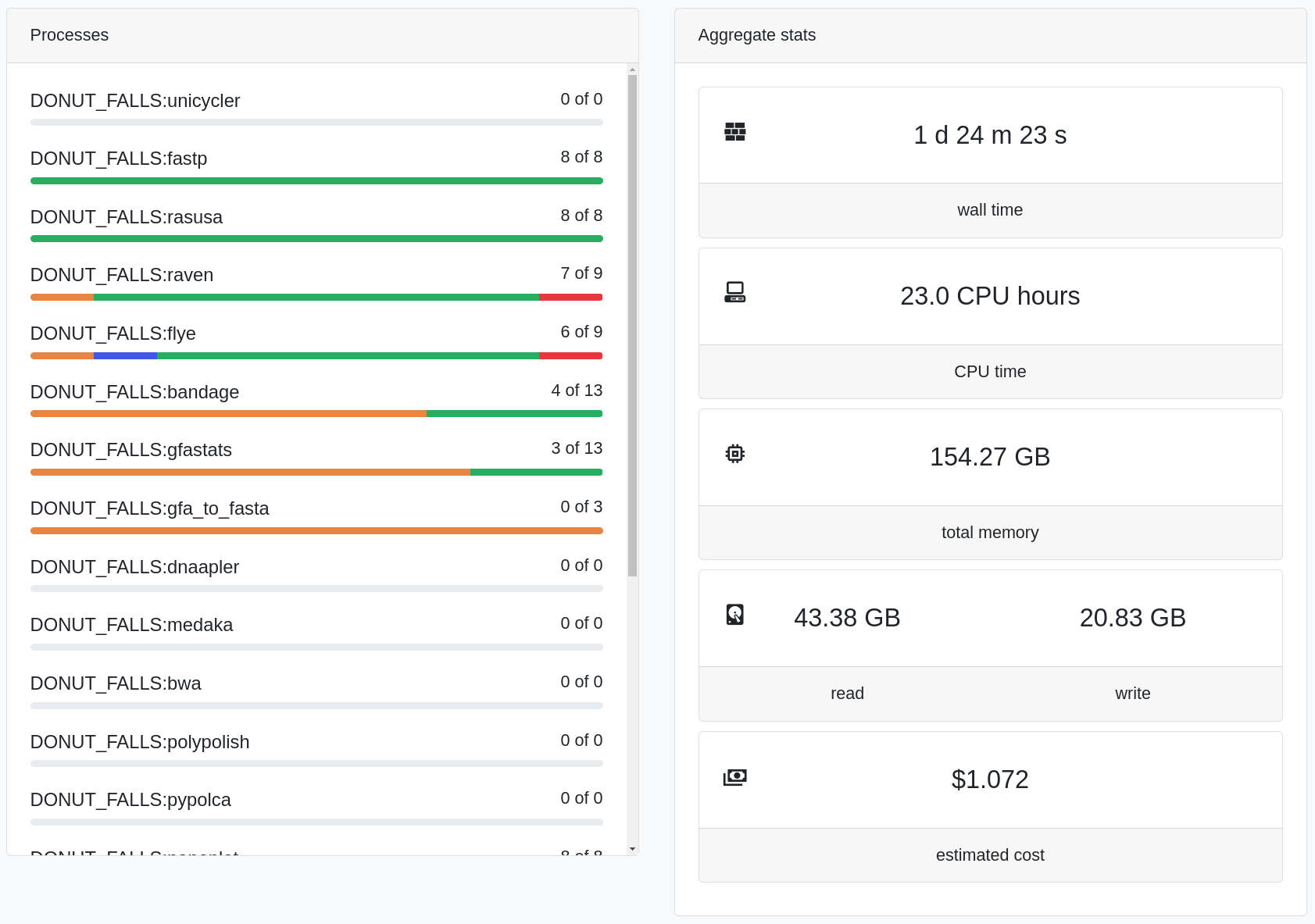
Monitoring Data Workflows with Nextflow Tower
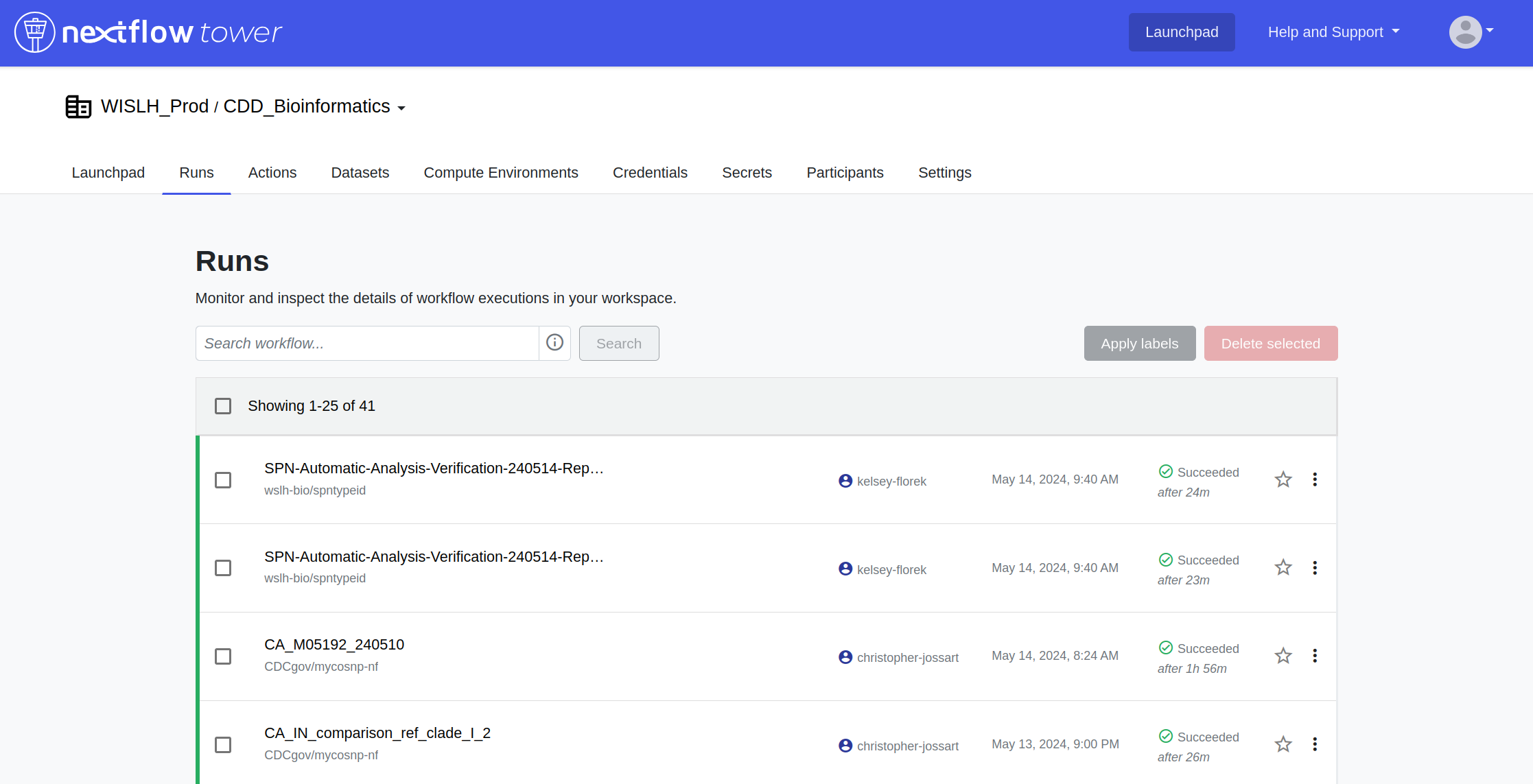
Monitoring Data Workflows with Nextflow Tower
WSLH Data Portal Infrastructure
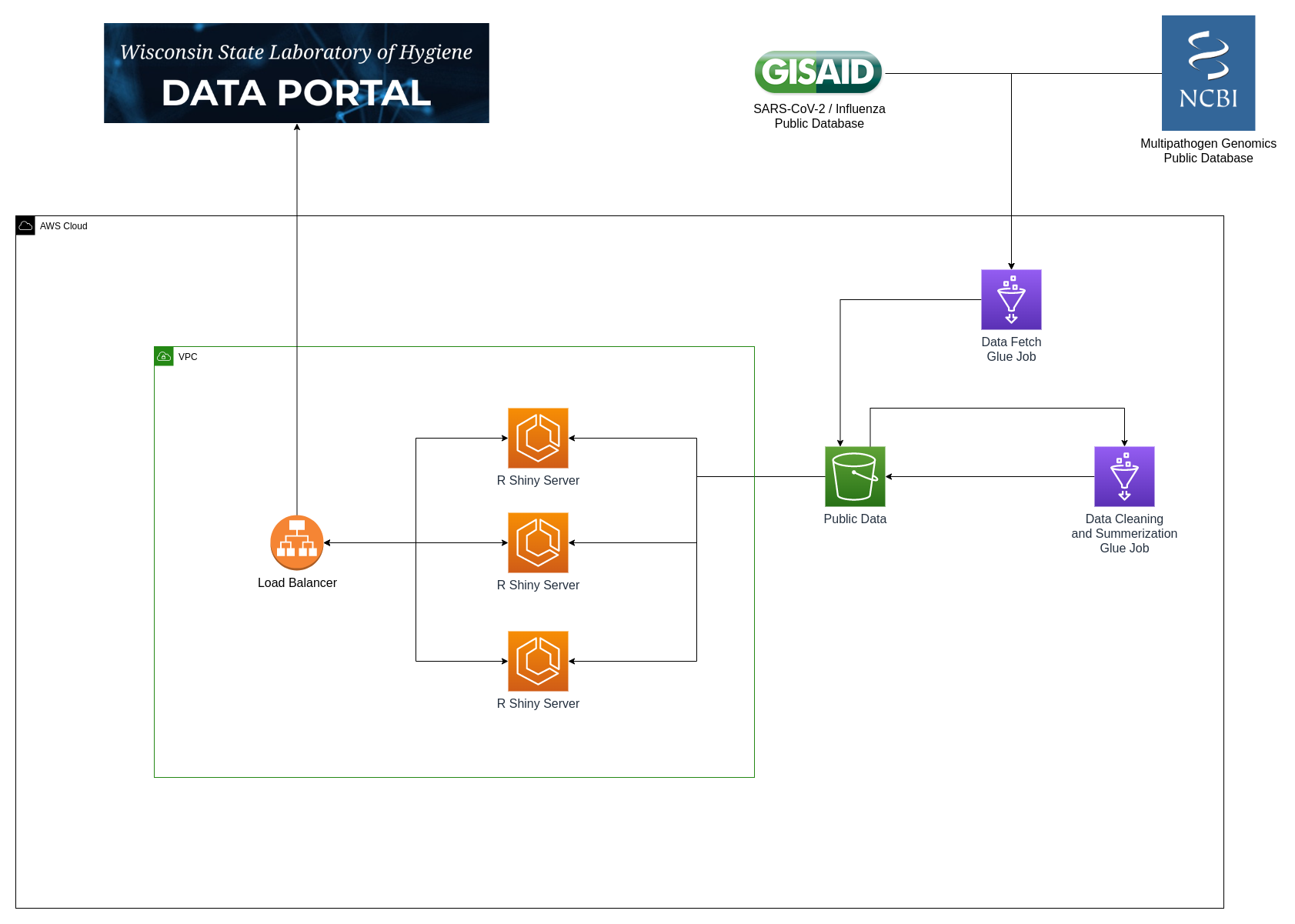
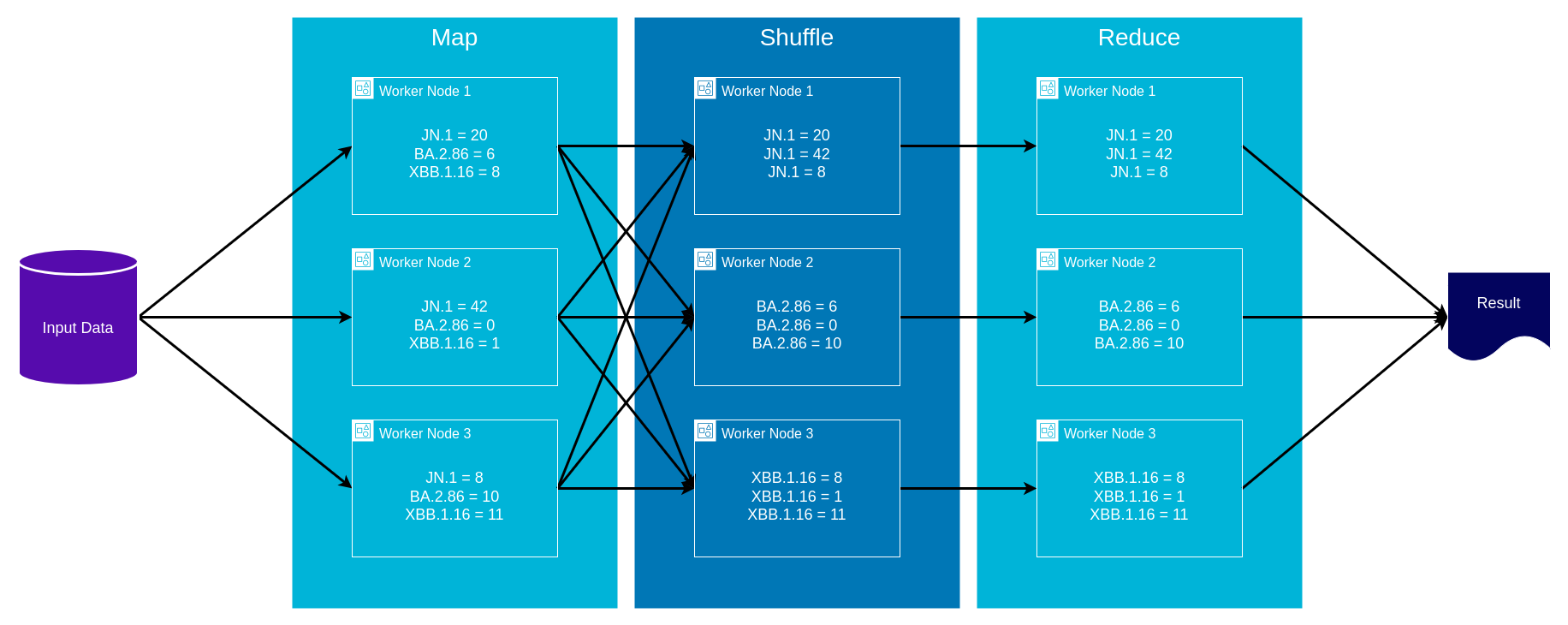
AWS Glue and MapReduce
- Map: input data is first split into smaller blocks, then each block is assigned to a separate worker node and processed in parallel
- Shuffle: worker nodes redistribute data such that all data belonging to a group located on the same worker node
- Reduce: if needed worker nodes then process each group of output data, in parallel
AWS Glue and MapReduce
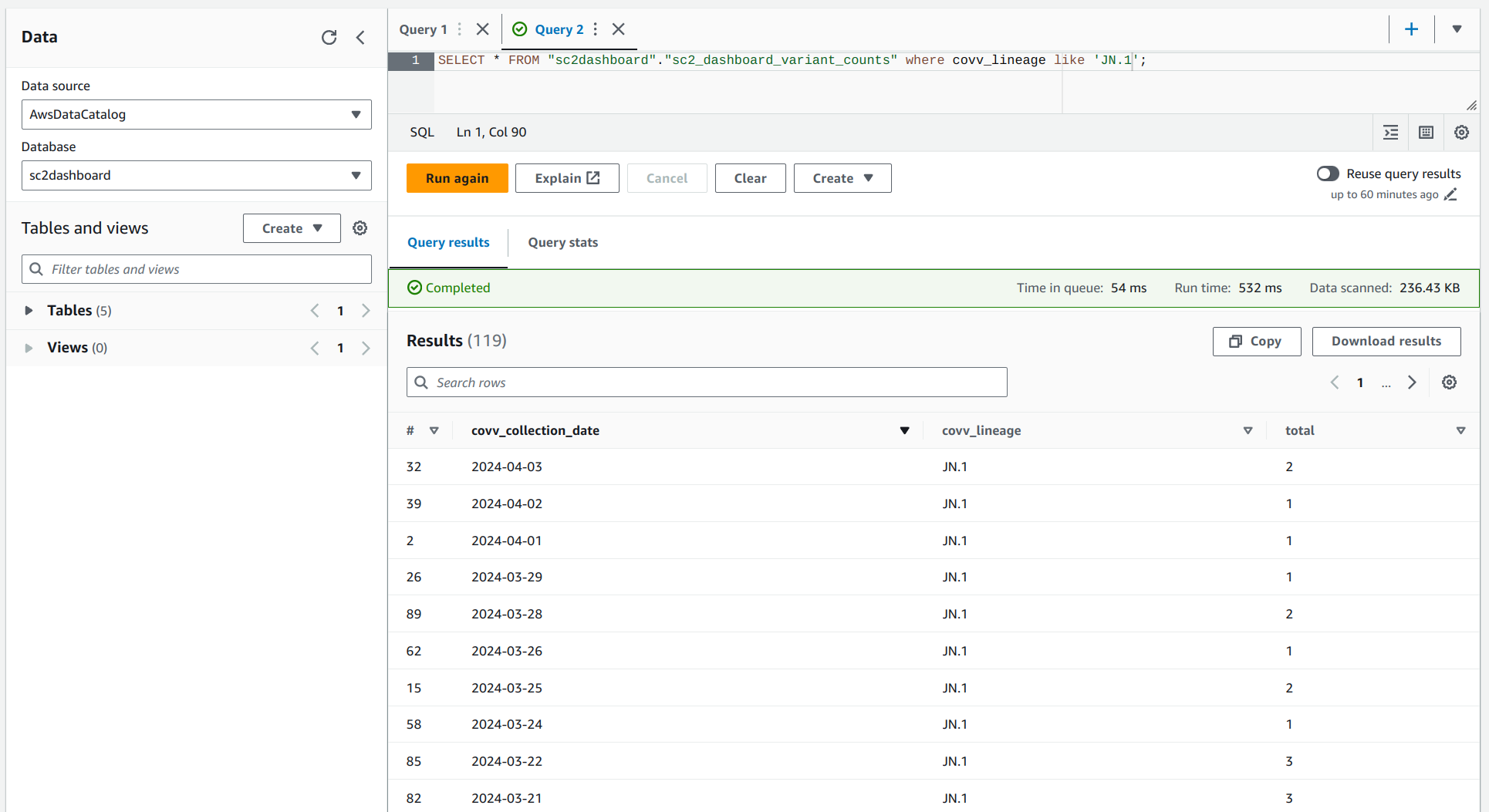
Query Data in the Data Lake
Simplifying Genomics for Public Health Partners
AMD Bioinformatics Regional Resource - Midwest Region
Ad-hoc Analytical Support
Provision of Computational Resources
Easy Genomics Partnership

Easy Genomics - Minimal Viable Product
- Simplify the process of launching and monitoring workflows
- Provide the ability for users to upload sequence data through the web browser
- Allow users to download analysis results through the web browser
- User/Lab separation
Easy Genomics - Sequence Data Upload

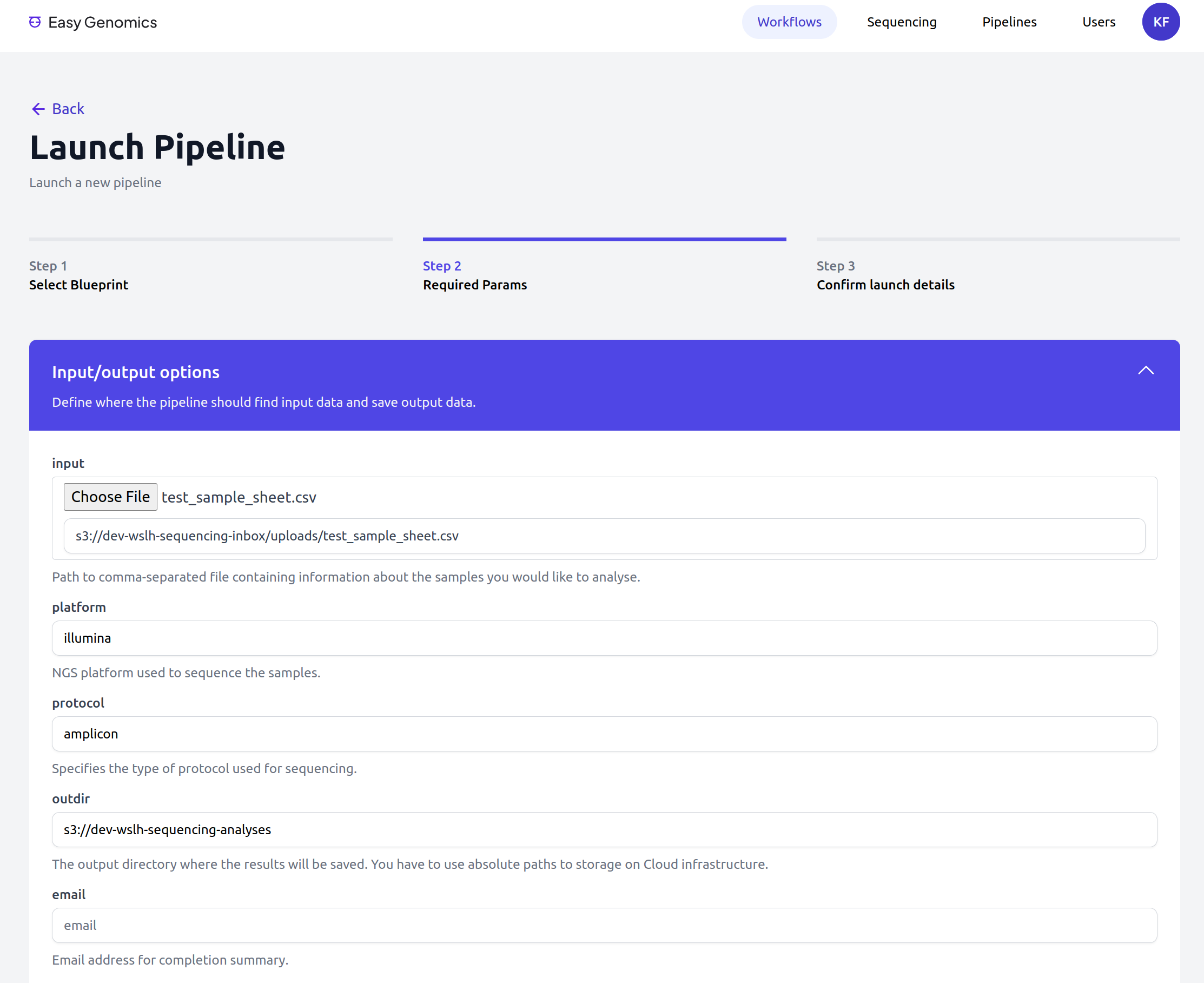
Easy Genomics - Launch
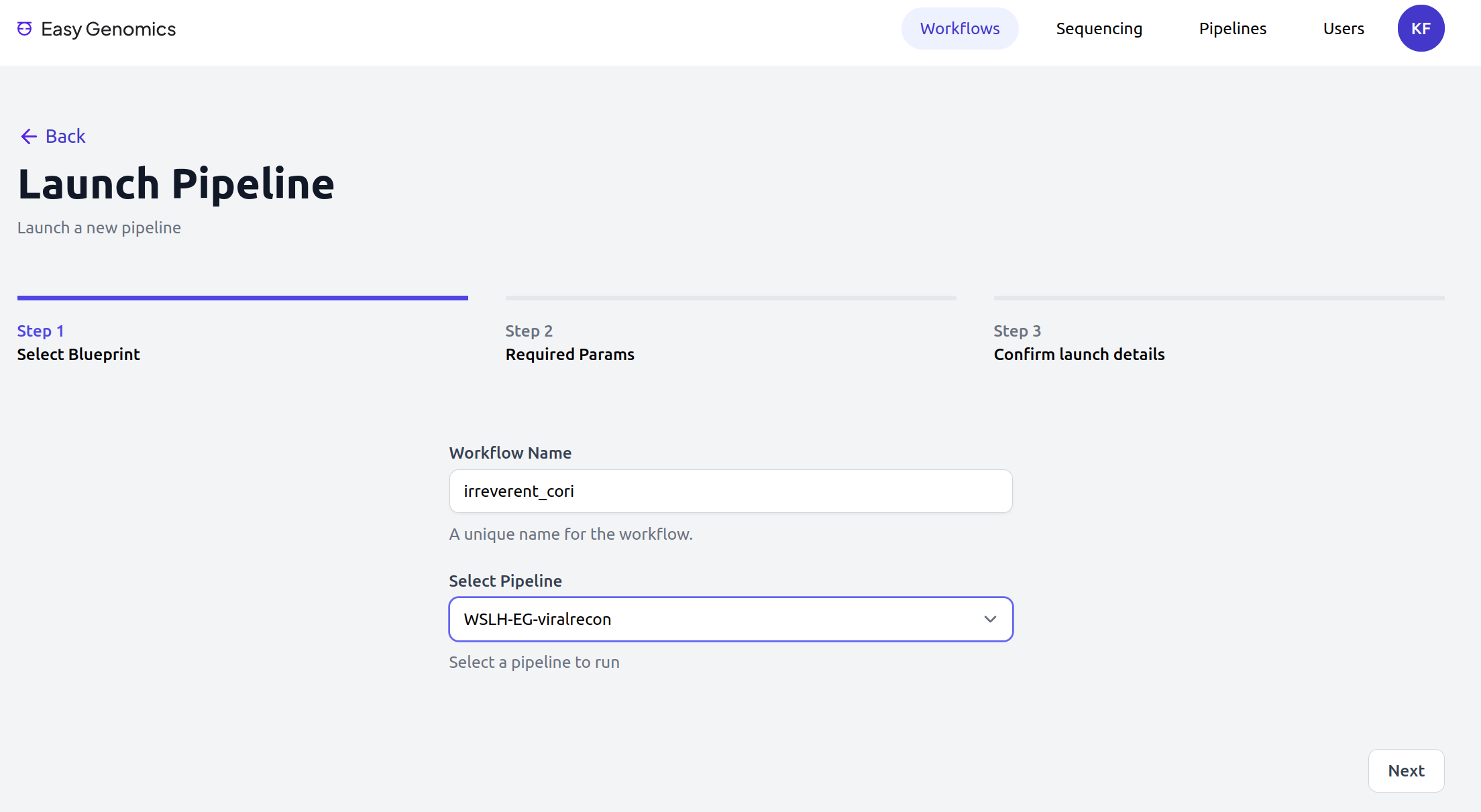
Easy Genomics - Launch
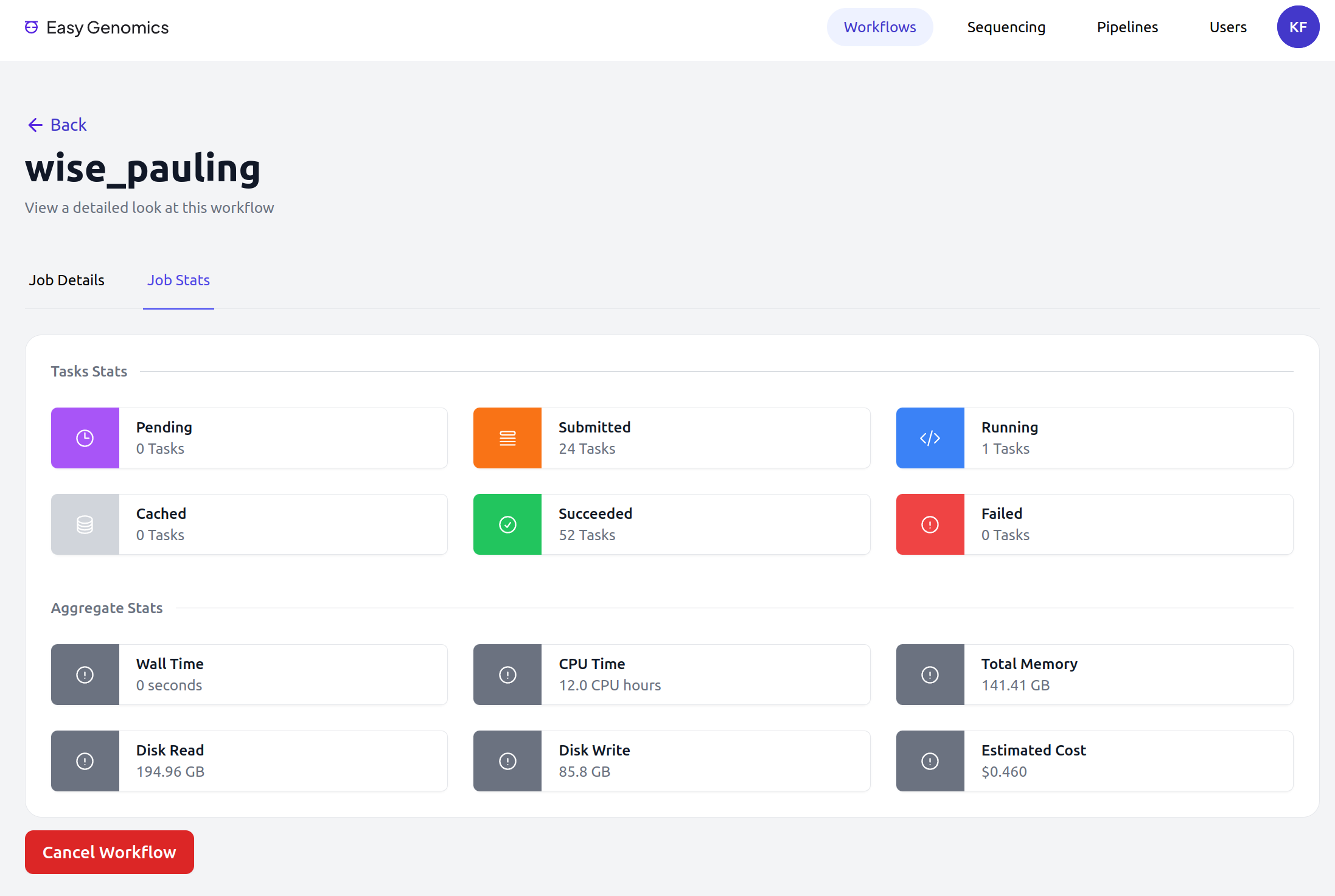
Easy Genomics - Monitor

Easy Genomics - Monitor
Easy Genomics - Roadmap
- 2024 Spring - Deploy Easy Genomics for internal use
- 2024 Early Summer - Open Access to SARS-CoV-2 Sequencing Laboratories
- 2024 Mid Summer - Easy Genomics MVP Update
- 2025 Winter - Deploy Easy Genomics in CDC National Bioinformatics Platform